Best DICOM Annotation Tools in 2026
TL;DR
A capable DICOM annotation tool helps medical AI teams do four things well:
- Label volumetric studies properly: CT and MRI require fast slice navigation, multi-planar viewing, and masks that stay consistent across slices for training stability.
- Run clinical QA at scale: maker-checker review, disagreement resolution, and slice-level traceability so labels are defensible in validation and audits.
- Keep datasets reproducible: versioned datasets and versioned label schemas so “model v2 vs v3” comparisons are not polluted by silent label drift.
- Clear security and governance hurdles: role-based access and logging that security teams typically require before PHI-bearing datasets are approved for use.
DICOM annotation tools covered in this blog:
- Taskmonk
- Kili
- V7
- Labelbox
- Encord
Introduction
A correct or incorrect medical diagnosis impacts treatment, care plans, and outcomes. As a result, computer vision and machine learning systems in medical imaging increasingly carry clinical and regulatory risk, not just technical complexity.
As most teams discover, it all starts with data. Getting a radiology AI product to market and building the evidence you need for serious clinical adoption depends on two things: data quality and speed.
Both are shaped by one practical layer that is often underestimated early on: how you annotate medical data(DICOM images) at scale, with consistent clinical rules and reviewability across CT, X-ray, PET, ultrasound, and MRI.
This guide is for medical AI teams and imaging orgs where DICOM annotation quality directly impacts model safety, validation, and time-to-deployment.
What is DICOM?
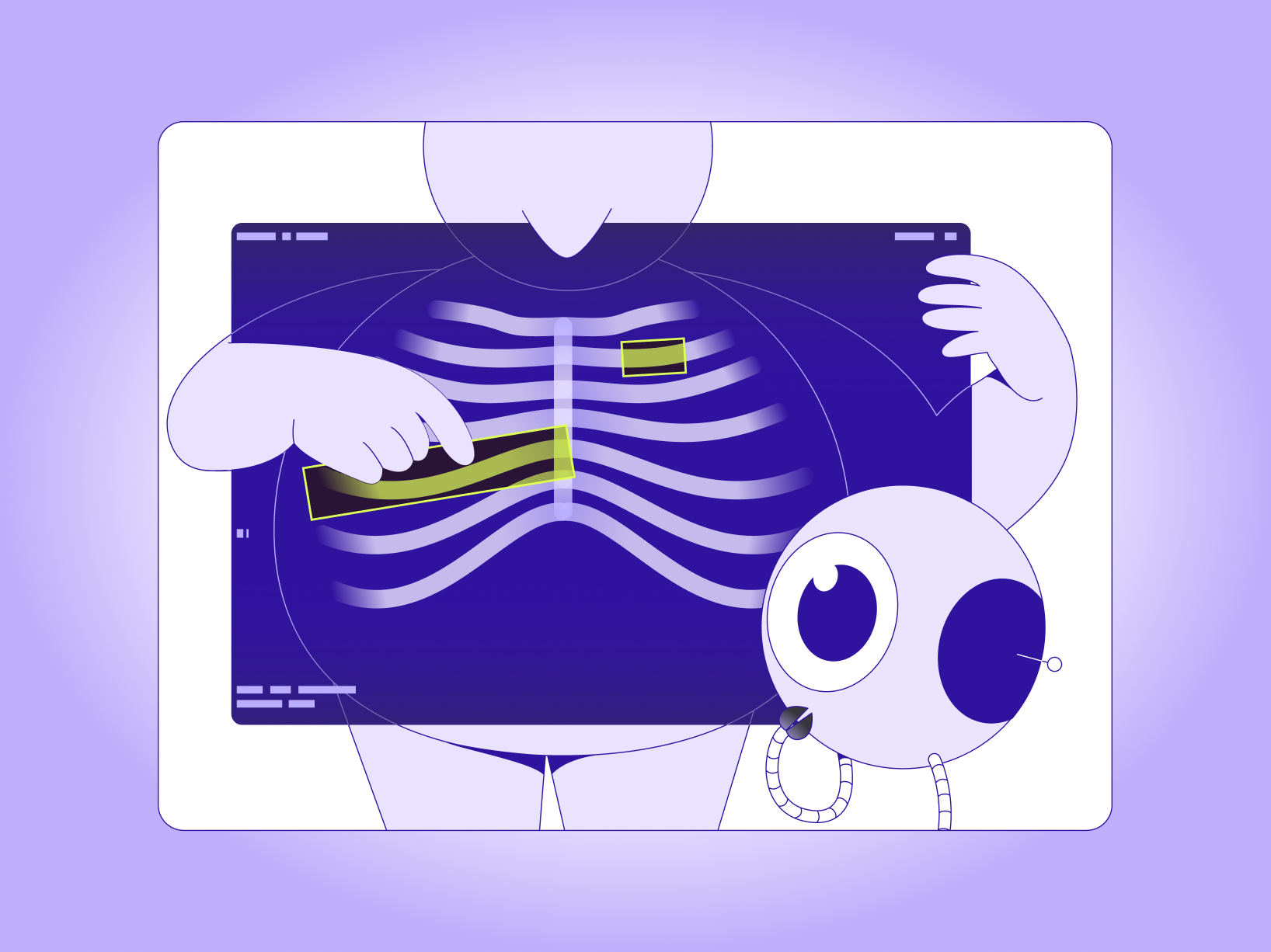
DICOM (Digital Imaging and Communications in Medicine) is the global standard for storing and exchanging medical images and their associated metadata. In plain terms, DICOM is not just a picture. It is typically a study made up of series, slices, acquisition context, and metadata that affect viewing, measurements, and annotation.
Because of that structure, DICOM annotation is not the same as labeling an ordinary image like a flat JPG or PNG. You are often annotating across slices within a study, preserving clinical context and measurement integrity so the labels remain useful for training and evaluation.
Head: Read more on DICOM annotation here.
How we compared these tools
Most DICOM annotation tools look “fine” in a short demo. The differences show up once you run real workloads with CT/ MRI/X-Ray volumes, multiple reviewers, evolving label rules, and repeated exports for model iterations.
We compared tools on the criteria that usually decide whether a medical imaging program scales cleanly or turns into rework.
- DICOM workflow fit: supports studies with multiple series, radiology navigation, window and level controls, and reliable measurements
- 3D and volumetric support: fast slice navigation, multi-planar viewing, consistent masks across slices, and exports that work for training and evaluation
- Annotation and label governance: supports your required annotation types, and keeps label schemas and instructions consistent as teams scale
- QA and clinical review: maker-checker review, disagreement handling, slice-level comments, plus basic metrics like rework and turnaround time
- Security and PHI readiness: role-based access, audit logs, and practical support for de-identification workflows
- Production fit: collaboration for radiologists and ops, integrations into data pipelines, and visibility into throughput vs rework
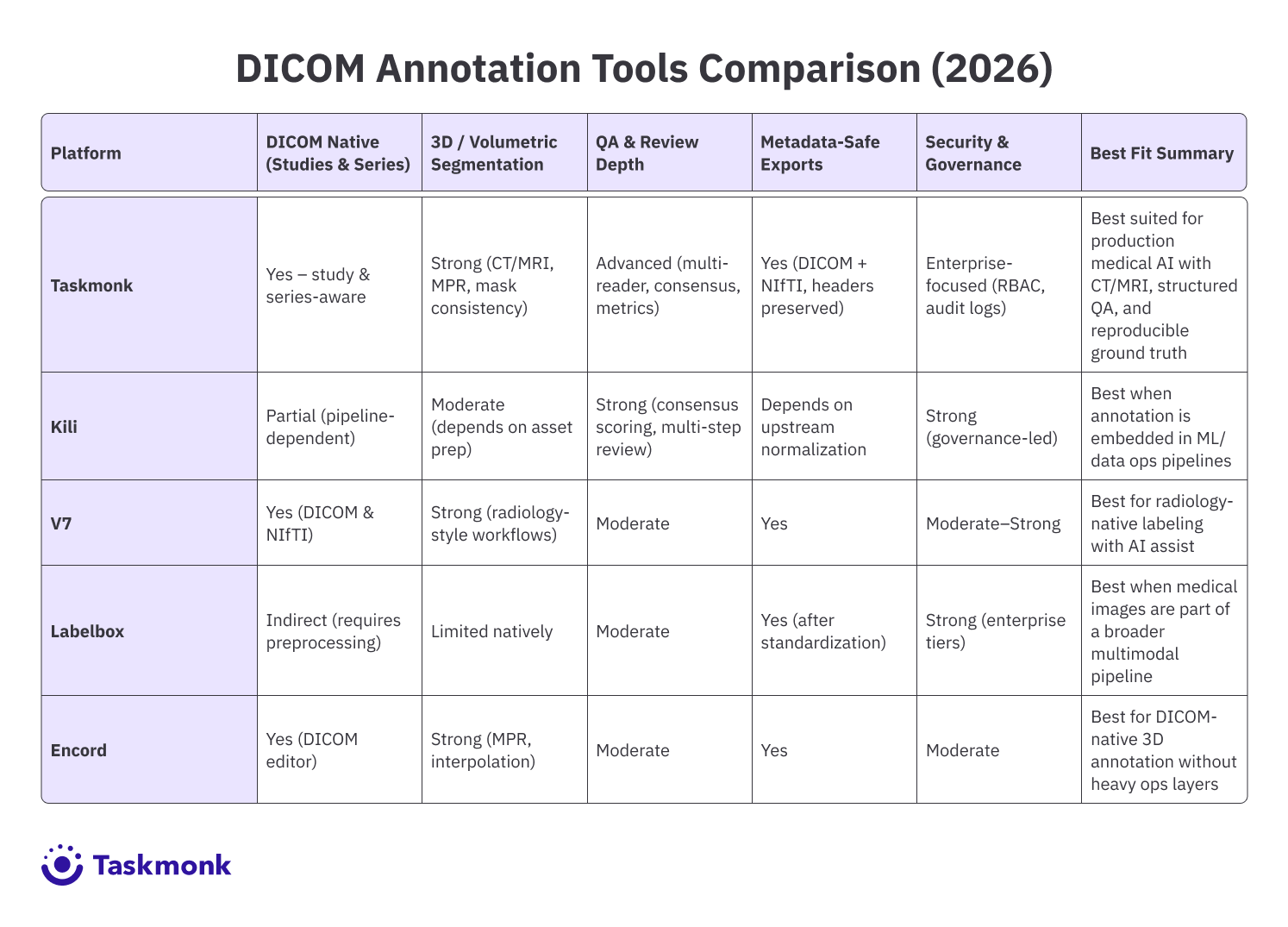
Best DICOM annotation tools
-
Taskmonk
Taskmonk is a DICOM annotation platform built for medical AI teams that need to operationalize DICOM annotation with predictable throughput, especially when workflows include CT and MRI volumes, multi-reader QA, and secure handling of PHI.
It supports DICOM series and NIfTI volumes, keeps critical metadata intact, and combines tooling with optional managed labeling delivery.
If your DICOM pipeline includes CT and MRI volumes, the tool choice shows up fast in rework and review time.
Taskmonk focuses on areas that usually break first at scale: volumetric navigation, consistent masks across slices, and structured QA, while keeping exports usable for repeated training and iteration.

Why Taskmonk stands out for DICOM annotation?-
Medical-format first, not “image-first.”
Imports DICOM series and NIfTI volumes while retaining headers such as UIDs, orientation, and pixel spacing. Exports preserve metadata and registration so masks align correctly across planes and downstream pipelines. -
Volumetric viewing plus radiology-grade tools
Supports window and level adjustments, MPR and MIP, and cine playback. Annotation tools include brush, polygon, spline, callipers, angle, and text, which cover common radiology labeling and measurement workflows. -
Automation that is tied to review, not just speed
Includes AI-assisted pre-labeling, mask propagation, and auto-segmentation, with low-confidence cases routed to expert review. Workflow routing and scheduled exports are positioned as part of keeping studies moving without bottlenecks. -
QA workflows designed for multi-reader programs
Built-in review steps, consensus reviews, and tracking of inter-reader variability are explicitly supported. Dashboards track speed, quality, and backlog so teams can scale without losing control of rework. -
Security and governance for PHI workflows
The platform states alignment with SOC 2, HIPAA, and GDPR, and lists controls like encryption, SSO, RBAC, audit logs, and optional VPC deployment. -
Documented setup for DICOM projects
Taskmonk’s documentation shows DICOM explicitly explaining the DICOM toolset and interaction modes inside the viewer, creating projects, pulling tasks, and validating that annotators are seeing studies correctly inside the task queue, etc.
Best for:
Medical AI teams running CT and MRI DICOM annotation with structured review and consensus QA, especially when they also need metadata-safe exports and reproducible ground truth across model iterations.
Not ideal for:
Small research teams that only need a lightweight local viewer and do not need customized DICOMworkflow, QA, or governance features
Rated G2 reviewers repeatedly call out ease of onboarding and day-to-day usability, plus quality controls and responsive support.
One review mentions quick setup and being operational within days, and another highlights reduced rework through collaboration and feedback loops. -
Medical-format first, not “image-first.”
-
Kili
Kili is a good fit if your team prefers to run DICOM annotation as part of a controlled data & MLpipeline, where data and ML ops handle ingestion and normalization, and labeling teams work on standardized assets with consistent metadata.
Medical AI teams consider it when DICOM annotation needs to scale across multiple labelers and reviewers with measurable quality controls.
It is less “radiology viewer first” and more “pipeline and governance first”, which can be a real advantage if your data team already controls how DICOM studies are de-identified, curated, and prepared before labeling.
Kili publicly lists a Free Trial that includes 200 assets and then moves into quote-based tiers.
Why Kili stands out for DICOM annotation-
Quality controls that are measurable, not subjective
Kili makes agreement a first-class concept. Consensus workflows allow multiple labelers to annotate the same asset and compute agreement scores, which supports multi-reader validation. -
Multi-step review that fits maker-checker operations
Multi-step review supports multiple validation levels across reviewer groups. This enables predictable routing, separation of duties, and repeatable quality gates as volume grows. -
Integration-friendly for ML stacks
Kili is also noted as an easy-to-integrate ML tool, which can be a practical fit when your labeling layer must plug into an existing medical data pipeline. -
Ontology and project lifecycle documentation
Kili’s setup flow covers ontology creation, role assignment, quality configuration, review, KPI analysis, and export, making it easier to operationalize projects consistently.
Best for:
Medical AI teams that want DICOM annotation governed through strong QA measurement, multi-step review, and operational controls.
Not ideal for:
Teams that want a radiology-native, study-first DICOM viewer experience with minimal upstream preparation and minimal workflow configuration. -
Quality controls that are measurable, not subjective
-
V7
V7 Darwin is one of the more radiology-native options in this list. It supports DICOM and NIfTI directly, and its medical imaging experience is designed to feel closer to a DICOM viewer than a generic labeling tool.
That makes it easier for annotation and review teams to work with little format workarounds.
Why V7 stands out for DICOM annotation
-
Radiology-focused tooling for day-to-day labeling speed
V7 documents workflow helpers that matter in real DICOM work, such as measurement tools and window-level controls, which reduce back-and-forth between the viewer and labeling steps. -
AI assist for medical imaging.
Auto Annotate is supported in DICOM and NIfTI workflows, with guidance on where it performs well and where manual tools, such as a brush, are needed. -
Volumetric handling
V7 highlights MPR-style workflows and volumetric file management, which helps when teams move from small pilots to consistent 3D segmentation programs.
Best for:
Medical AI teams that want a DICOM and NIfTI-native workflow with radiology-style controls and AI assist for faster throughput.
Not ideal for:
Teams that need a simple fixed monthly price, since usage-based pricing can require tighter monitoring as volumes grow. -
Radiology-focused tooling for day-to-day labeling speed
-
Labelbox
Labelbox is a flexible training data platform that is widely adopted for multimodal labeling and dataset operations.
For medical imaging, it positions its tooling around labeling medical imagery, such as pathology scans, images, and videos, with workflow support that helps teams iterate faster once data is organized inside the platform.
Labelbox pricing is usage-based through Labelbox Units (LBUs), with Free, Starter, and Enterprise tiers.
Why Labelbox stands out for DICOM annotation?
-
Strong “data engine” workflow for continuous iteration
Labelbox is built around dataset operations, workflow orchestration, and model-driven loops, which help when annotation is ongoing and tied to model experimentation, not one-time labeling. -
Flexible annotation stack once assets are standardized
If your pipeline standardizes DICOM into labelable assets and consistent metadata, Labelbox becomes useful for managing labeling at scale across teams and tasks. -
Predictable cost modeling for ops teams
The LBU model makes it easier to forecast annotation costs based on volume and workflow usage, especially as volumes ramp up.
Best for:
Teams that want a mature training data platform for workflow operations and model iteration, and already have a pipeline to standardize medical imaging assets for labeling.
Not ideal for:
Teams looking for a radiology-first, study-native DICOM experience, especially for heavy CT and MRI volumetric programs -
Strong “data engine” workflow for continuous iteration
-
Encord
Encord offers a dedicated DICOM editor with features aimed at CT, X-ray, and MRI labeling, including multi-planar workflows, DICOM metadata visibility, and slice-to-slice tooling that supports 3D annotation consistency.
Why Encord stands out for DICOM annotation?
-
MPR and slice-linked annotation behavior for 3D work
Encord’s DICOM editor supports toggling between axial, coronal, and sagittal views, and its MPR approach is designed so annotations carry across views for consistent anatomy labeling. -
Radiology-grade viewing controls and metadata visibility
It supports custom window widths and levels, natively displays DICOM metadata, and is built to handle higher pixel intensity ranges expected in medical imaging workflows. -
3D workflow accelerators that reduce manual rework
The DICOM editor includes object tracking and slice interpolation, plus automation features to reduce manual segmentation effort.
Best for:
Medical AI teams that want a DICOM-native platform with strong 3D workflows, radiology controls, and tooling that supports repeatable exports across model iterations.
Not ideal for:
Small teams that want a lightweight, standalone tool with no platform overhead, without collaboration, governance, or auditability. -
MPR and slice-linked annotation behavior for 3D work
A few more Open Source DICOM annotation tools to consider
Open source tools are often a good fit for research, prototyping, and early feasibility work, especially when you need full control of the stack more than operational scalability.
-
3D Slicer
3D Slicer is a free, open-source platform used for medical image visualization and segmentation, with explicit DICOM support for importing, exporting, and even network transfer in clinical research workflows. It has a large ecosystem of extensions, which is why it is common in academic pipelines where teams need custom modules or repeatable research workflows.
Best for: research-grade CT and MRI segmentation, prototyping new labeling protocols, and teams that want flexibility over enterprise workflow features. -
ITK-SNAP
ITK-SNAP is a focused open-source tool built around 3D and 4D image segmentation, and it supports formats commonly used in medical imaging workflows, including DICOM and NIfTI. It is popular when the main requirement is fast, reliable segmentation and manual correction workflows without needing a full annotation operations layer.
Best for: researchers and small teams doing 3D segmentation on CT and MRI, where you want a straightforward segmentation-first workflow. -
MITK Workbench
MITK Workbench is a free application built on the Medical Imaging Interaction Toolkit (MITK) and designed for viewing, processing, and segmenting medical images. It is widely used in medical image processing research, where teams may also build or extend interactive imaging applications on top of MITK.
Best for: teams that want an open-source workstation plus a developer-friendly toolkit for interactive medical imaging workflows. -
OHIF Viewer
OHIF is an open-source, browser-based medical imaging viewer that supports modern workflows like DICOMweb and uses Cornerstone3D for decoding, rendering, and annotation. Because it is web-native and extensible, it is often used as a base for custom clinical viewing workflows and lightweight labeling experiences, especially when data is already accessible through DICOMweb-compatible archives.
Best for: web-based DICOM viewing and annotation workflows, teams building custom imaging apps, and organizations standardizing on DICOMweb.
Conclusion
A significant indicator of where medical imaging is headed is DICOM’s push toward web-native interoperability.
In November 2025, the DICOM Standards Committee circulated “DICOMweb Send” (Supplement 248) for public comment, extending the way imaging studies can be moved and shared through modern web workflows.
At the same time, major industry forums like RSNA 2025 continued to spotlight DICOMweb and FHIR as the backbone of connected, cloud-ready imaging ecosystems.
That shift matters because annotation is no longer happening on isolated folders of scans. It increasingly sits inside a pipeline where teams need to ingest studies cleanly, de-identify where required, coordinate expert review, and export ground truth in formats that survive model iteration.
Head: Build review-ready ground truth for medical imaging. Explore how Taskmonk helps medical AI teams
FAQs
-
What are DICOM annotation tools used for?
DICOM annotation tools help teams label medical imaging studies so they can train and validate medical AI models. Typical outputs include segmentation masks, bounding boxes, keypoints, and measurements used for tasks like lesion detection, organ segmentation, triage prioritization, and quantitative imaging. -
Which DICOM annotation tools are best for medical AI projects?
For production medical AI, teams usually prioritize tools that support volumetric workflows, structured QA, and reproducible exports. In this guide, we covered five widely shortlisted platforms: Taskmonk, Kili, V7, Labelbox, and Encord. The best choice depends on whether your biggest constraint is 3D labeling depth, clinical review bandwidth, or pipeline integration. -
Do DICOM annotation platforms need to be HIPAA compliant?
If your dataset contains PHI and you operate as a covered entity or business associate in the US, you typically need HIPAA-aligned controls and a Business Associate Agreement (BAA) with the vendor. In practice, teams also look for role-based access, audit logs, encryption, and clear de-identification workflows. -
Can DICOM annotation tools handle 3D medical imaging and volumetric data?
Yes, but capability varies a lot. For CT and MRI, look for slice-aware segmentation, multi-planar navigation, interpolation tools, and exports that preserve spacing and orientation so masks remain valid in 3D training and evaluation. -
What export formats should a DICOM annotation tool support for medical AI?
At a minimum, you want exports that fit your pipeline without custom rework. Common requirements include DICOM RTSTRUCT (in some radiotherapy workflows), NIfTI masks for 3D segmentation training, and JSON-based exports for project metadata and review traces. The right answer depends on whether you train 2D slice models, 3D volumes, or hybrid approaches. -
How do medical AI teams avoid label drift across annotators and sites?
Use a calibration set early, define a strict labeling protocol, and enforce maker-checker review with measurable QA signals like rework rate and agreement. If you add new scanners or new sites, re-run calibration because protocol differences can shift how anatomy and pathology appear, which changes labeling decisions.
.webp)